a. Go to File>Import>Network from Table (Text/MS Excel)... |
b. Go to Select File(s) and locate AutoSOME output file (4) containing all edges (see Files written to disk in the manual) or use the tutorial dataset. |
c. Set Source Interaction to Column 1 and Target Interaction to Column 2. Finally, click on Column 3 in the data Preview window to activate it (it will turn blue). |
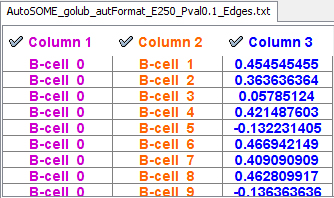 |
d. Select Import and then Close. A raw network will appear as a grid.
|
 |
e. Go to File>Import>Attribute from Table (Text/MS Excel)...
|
f. Go to Select File(s) and locate AutoSOME output file (5) containing all nodes and attributes (see Files written to disk in the manual) or use the tutorial dataset. |
g. Select Import. |
h. Change global properties of the network: |
|
| |
i. In the VizMapperTM tab:
|
j. Next to Node Label, click ID and select Column 3. All nodes should now be relabeled according to the original data labels. Minimize the Node Label property by selecting the minus icon. |
k. Under Unused Properties find Edge Color and double-click it. |
|
| |
l. Find Edge Line Width under Unused Properties and double-click it.
|
|
| |
m. Find Edge Opacity under Unused Properties and double-click it.
|
|
| |
n. Finally, find Node Color under Unused Properties and double-click it.
|
|
| |
o. Go to Layout in the main menu and select Settings
|
|
| |
p. To export the final network, go to File>Export>Network View as Graphics...
|
q. Select file format and save image! |
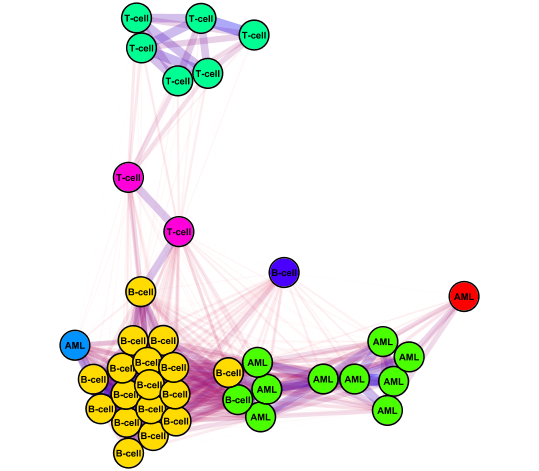 |
r. To expedite this process for next time, start with your saved network. All visualization parameters will still be specified. Then, simply input your edge and node files and perform the layout. Note that for the Node Color property, it is important to assign colors again using the process given in step (n). |



